DNA Rotator Walk

Requires a Wolfram Notebook System
Interact on desktop, mobile and cloud with the free Wolfram Player or other Wolfram Language products.
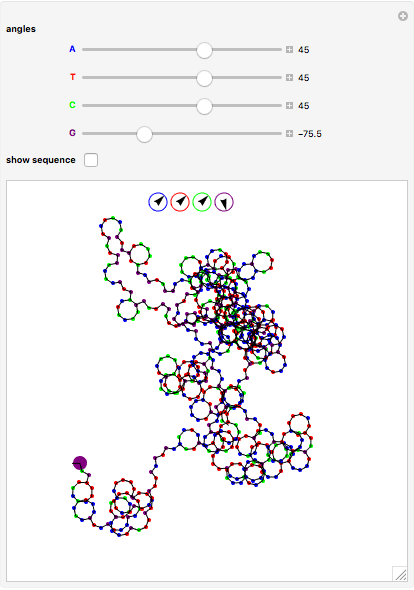
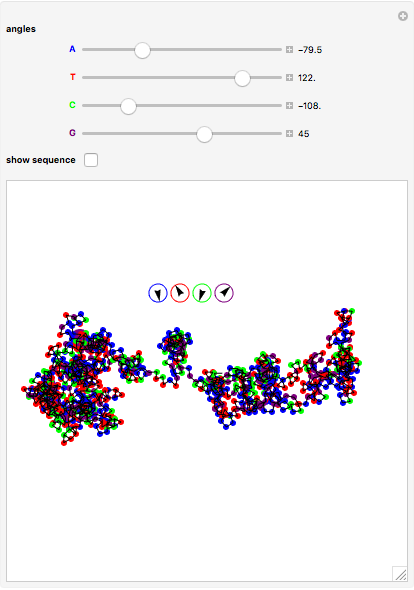
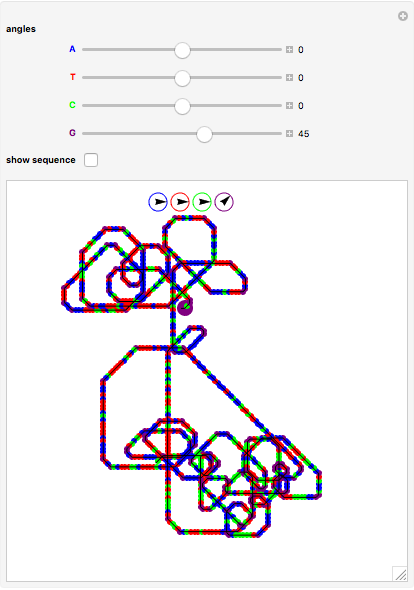
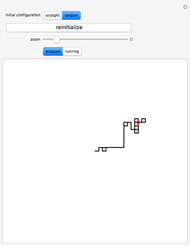
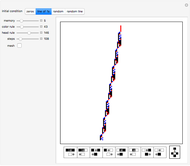
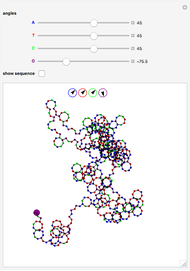
The DNA rotator walk works by reading the DNA sequence one base at a time ( ,
,  ,
,  , or
, or  ), turning through an angle that depends on the DNA base, and moving ahead by one step. For example, one might choose
), turning through an angle that depends on the DNA base, and moving ahead by one step. For example, one might choose  for
for  (clockwise),
(clockwise), 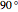 for
for  (counterclockwise),
(counterclockwise), 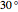 for
for  (counterclockwise), and
(counterclockwise), and  ° for
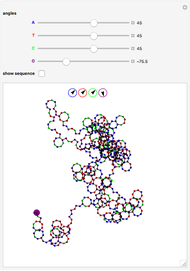
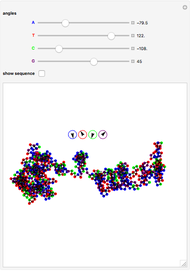
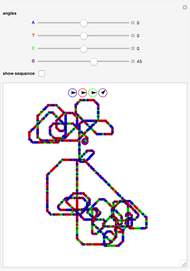
° for  (clockwise). The directions chosen for each base are indicated by the colored arrows at the top of the graphic. The starting point is indicated by a larger dot, and the starting direction is always upward. The DNA sequence used here is a 1000-bp region taken from a chicken gene called embryonic skeletal MHC. The gene region includes an exon surrounded by two introns. This visualization is useful to get an intuition for the properties of a DNA sequence. For example, with appropriate choices of angles, long repeat sequences such as poly-
(clockwise). The directions chosen for each base are indicated by the colored arrows at the top of the graphic. The starting point is indicated by a larger dot, and the starting direction is always upward. The DNA sequence used here is a 1000-bp region taken from a chicken gene called embryonic skeletal MHC. The gene region includes an exon surrounded by two introns. This visualization is useful to get an intuition for the properties of a DNA sequence. For example, with appropriate choices of angles, long repeat sequences such as poly-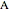 sequences ("
sequences (" ") will produce localized circles, and sequences with equal mixtures of bases will tend to produce randomized regions.
") will produce localized circles, and sequences with equal mixtures of bases will tend to produce randomized regions.
Contributed by: Paul-Jean Letourneau (March 2011)
Open content licensed under CC BY-NC-SA
Snapshots
Details
detailSectionParagraphPermanent Citation
"DNA Rotator Walk"
http://demonstrations.wolfram.com/DNARotatorWalk/
Wolfram Demonstrations Project
Published: March 7 2011