Boolean NK Networks

Requires a Wolfram Notebook System
Interact on desktop, mobile and cloud with the free Wolfram Player or other Wolfram Language products.
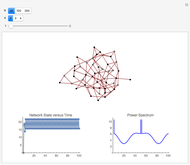
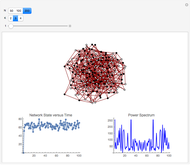
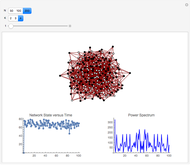
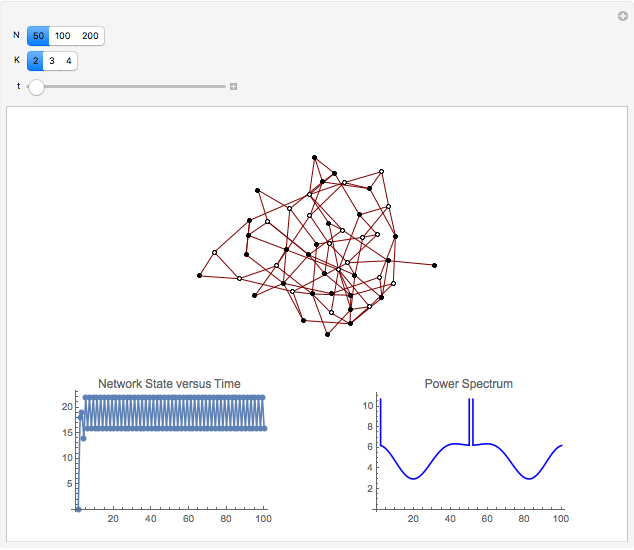
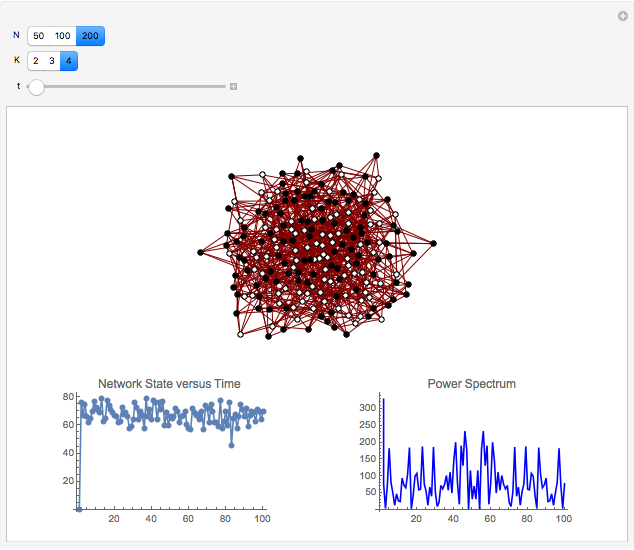
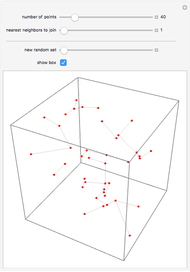
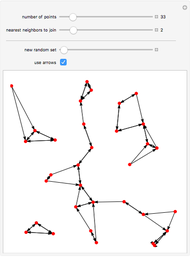
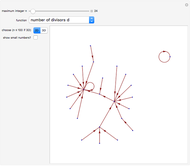
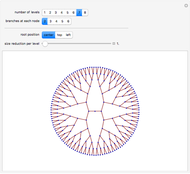
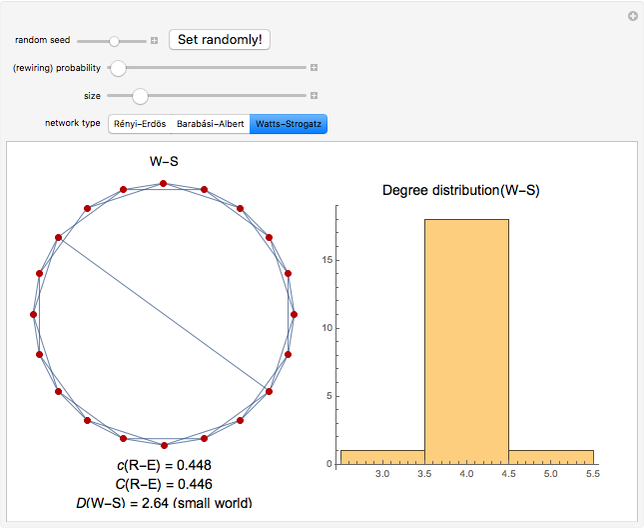
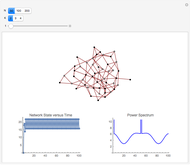
Boolean NK networks are networks whose nodes can be in one of two states (on or off) at any particular time in their dynamical evolution. The dynamics of Boolean NK networks are determined by two parameters ( , which defines the number of nodes in the network, and
, which defines the number of nodes in the network, and  , which defines the number of directional links between nodes) and a rule table for each node that specifies the state of the node at time step
, which defines the number of directional links between nodes) and a rule table for each node that specifies the state of the node at time step  given the states of the connecting nodes at time step
given the states of the connecting nodes at time step  . In this Demonstration, each node is randomly assigned both an initial state and a rule table. Different values of
. In this Demonstration, each node is randomly assigned both an initial state and a rule table. Different values of  and
and  result in different network dynamics, which can be seen in the plot of network state (defined as the Hamming distance between the network's present state and initial state) versus time and the power spectrum of this plot. By varying
result in different network dynamics, which can be seen in the plot of network state (defined as the Hamming distance between the network's present state and initial state) versus time and the power spectrum of this plot. By varying  , you can directly observe the real-time evolution of the network, which appears as a system of blinking lights.
, you can directly observe the real-time evolution of the network, which appears as a system of blinking lights.
Contributed by: Garrett Neske (March 2011)
Open content licensed under CC BY-NC-SA
Snapshots
Details
Boolean NK networks were introduced by theoretical biologist Stuart Kauffman as a model of genetic regulatory networks, with genes as nodes and Boolean states as gene expression or repression. A key discovery was that, with  , the network exhibited dynamical properties characteristic of biological systems, such as homeostasis and evolvability. While these properties are observed in a network of Boolean elements, they are not properties of the elements themselves: a phenomenon known as emergence or self-organization.
, the network exhibited dynamical properties characteristic of biological systems, such as homeostasis and evolvability. While these properties are observed in a network of Boolean elements, they are not properties of the elements themselves: a phenomenon known as emergence or self-organization.
Reference:
S. A. Kauffman, The Origins of Order: Self-Organization and Selection in Evolution, New York: Oxford University Press, 1993.
Permanent Citation