Antibody Production Fed-Batch

Requires a Wolfram Notebook System
Interact on desktop, mobile and cloud with the free Wolfram Player or other Wolfram Language products.
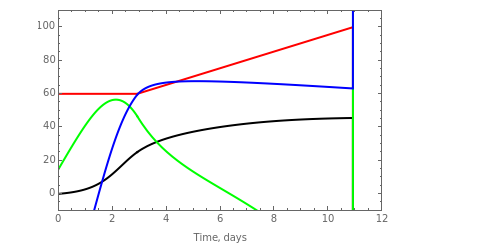
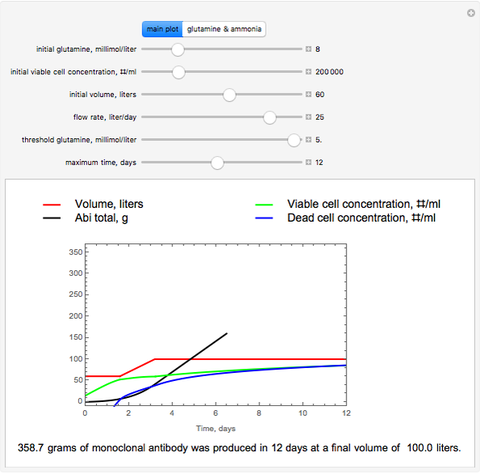
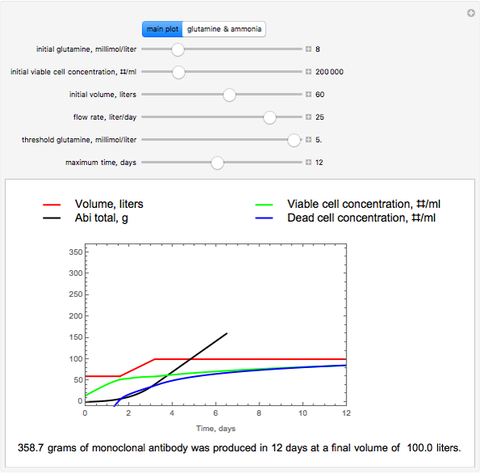
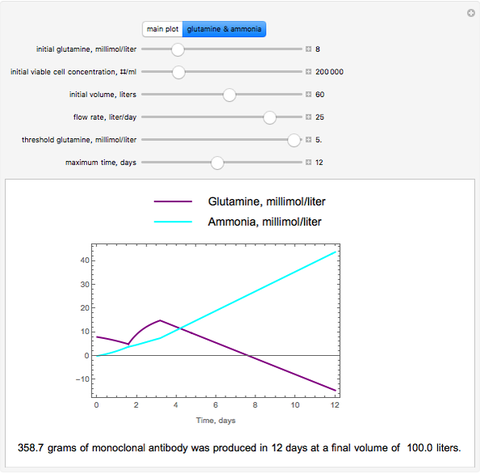
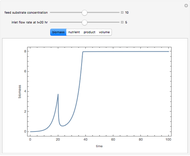
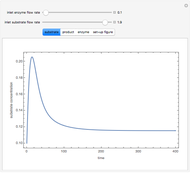
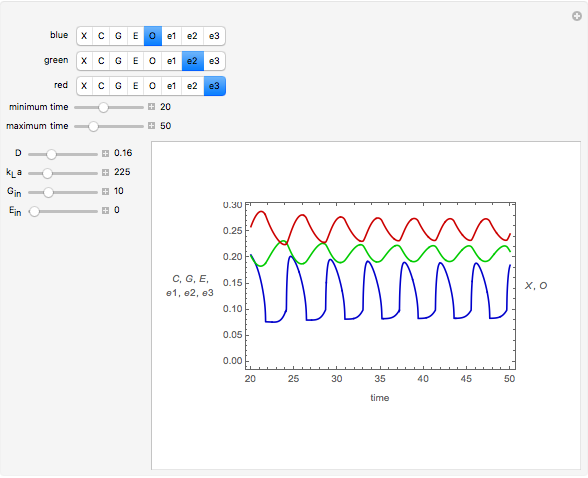
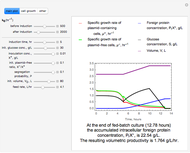
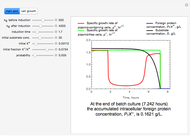
The dynamic profiles show the variation of different process variables in a typical fed-batch culture of hybridomas to produce monoclonal antibodies. The user can vary a number of process variables that a fed-batch operator can implement in a real fed-batch process, such as the initial volume, initial glutamine concentration, feeding rate, threshold concentration of glutamine at which feed is turned on, inoculum (initial viable cell concentration), and the duration of fed-batch culture.
[more]
Contributed by: David J. W. Simpson and Dhinakar S. Kompala (March 2011)
Open content licensed under CC BY-NC-SA
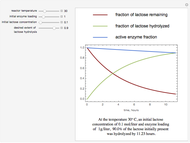
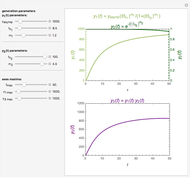
Snapshots
Details
The underlying kinetic model used in these interactive simulations is based on the model proposed by Glacken et al (1989), "Mathematical Descriptions of Hybridoma Culture Kinetics: 3. Simulation of Fed-Batch Bioreactors," Journal of Biotechnology, 10(1), 1989 pp. 39–65. One modification made to the published model in this simulation is the inclusion of an "inverse growth associated" production of monoclonal antibody as such production kinetics are more typically found in murine hybridoma cultures. In this dated work, glutamine is assumed to be the major limiting nutrient, and the other major limiting nutrient, glucose, is presumed to be in excess, which is a typical case. Ammonia, the metabolic by-product from glutamine metabolism, is toxic to cell growth and can trigger cell apoptosis if ammonia is accumulated in the fed-batch bioreactor at a high enough concentration. So adding too much glutamine at the beginning of a batch culture will reduce the production of viable cells and thereby the production of the desired product, monoclonal antibody.
In later developments however, recombinant Chinese hamster ovary cells are used for the fed-batch production of humanized antibodies. These CHO cells are also typically grown in fed-batch cultures, which need to be optimized for maximizing antibody titers by manipulating many of the same process variables shown above. One major improvement in newer cell lines is the specific productivity of the more recent cell lines. This model constant can be readily changed in the underlying model equations to reflect the typically obtained higher antibody titers (> 5g/l) in recent industrial practice. Some of these newer CHO cells have a glutamine synthetase (GS) gene as the selection marker and such cells do not need to be fed glutamine. Instead glutamate is converted inside the cells to synthesize glutamine. In such cells, glucose is likely to be the limiting nutrient and its metabolic by-product lactate is the toxic metabolite which could limit the cell growth at high lactate concentrations. The optimization of feeding rate is still needed to maximize the antibody accumulations at the end of a fed-batch culture by minimizing the lactate accumulation, which is quite analogous to the present simulation.
Here we explicitly give the differential equations being modeled (refer to the program for the values of constants used):
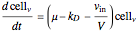 ,
,
 ,
,
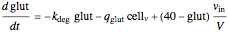 ,
,
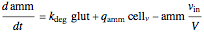 ,
,
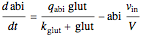 ,
,
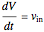 ,
,
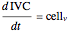 ,
,
where
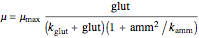 ,
,
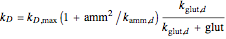 ,
,
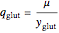 ,
,
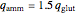 ,
,
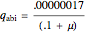 ,
,
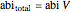 .
.
The value of 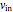 is fixed at zero during the initial batch phase. When the concentration of glutamine reaches a user-set threshold value, the concentrated nutrient feed (with high glutamine concentration of 40mM) is turned on and the flow rate
is fixed at zero during the initial batch phase. When the concentration of glutamine reaches a user-set threshold value, the concentrated nutrient feed (with high glutamine concentration of 40mM) is turned on and the flow rate 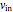 is set by the user on the slider. However once the culture volume,
is set by the user on the slider. However once the culture volume, 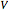 , has reached the maximum volume of the bioreactor (100 liters), nutrients can no longer be fed into the system; thus the value of
, has reached the maximum volume of the bioreactor (100 liters), nutrients can no longer be fed into the system; thus the value of 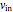 is reduced to zero and remains at this value for all later time. Consequently the concentration of glutamine may eventually fall below the user-specified threshold.
is reduced to zero and remains at this value for all later time. Consequently the concentration of glutamine may eventually fall below the user-specified threshold.
Permanent Citation