Network Motifs and Graphlets

Requires a Wolfram Notebook System
Interact on desktop, mobile and cloud with the free Wolfram Player or other Wolfram Language products.
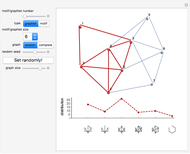
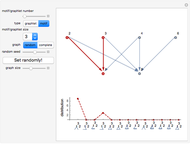
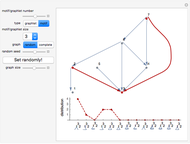
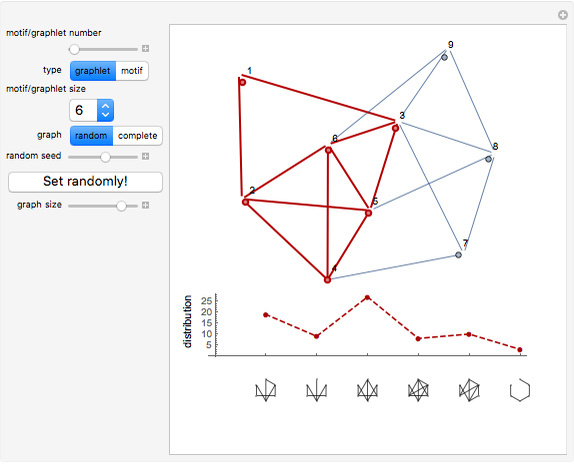
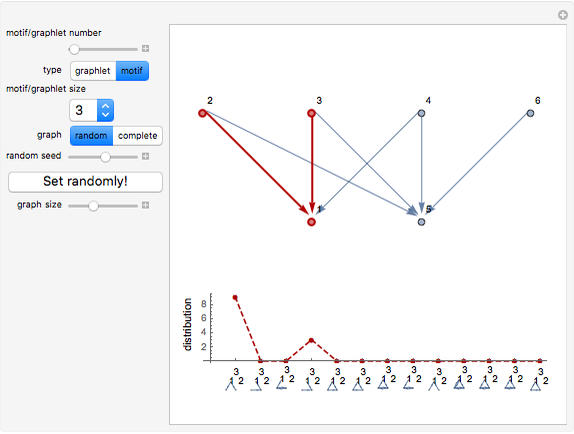
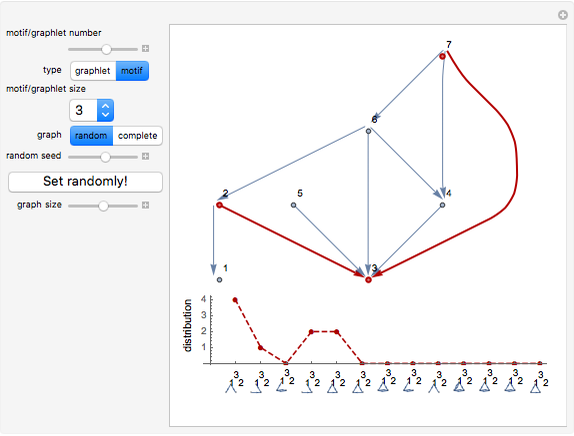
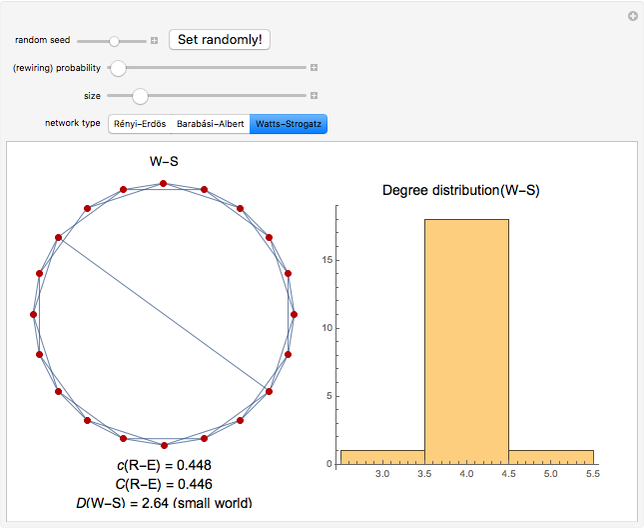
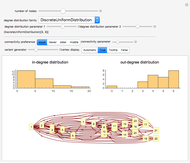
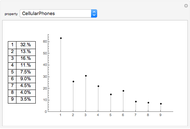
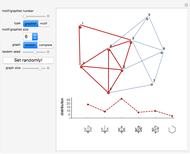
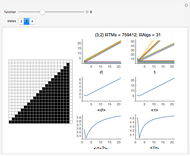
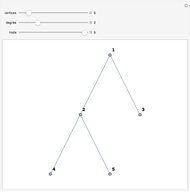
Network graphlets and motifs are statistically highly recurrent patterns in graphs and networks that have been found to characterize families of networks and shed light on biological functions under evolutionary selective pressure (when found in biological networks). Graphlets are undirected motifs. This Demonstration illustrates the motif and graphlet concept by highlighting all the subgraphs of a given size and plotting the distribution of these under the network in question. Finding motifs is very computationally expensive and so only motifs of size three are calculated in real time in this Demonstration.
Contributed by: Hector Zenil (December 2013)
Open content licensed under CC BY-NC-SA
Snapshots
Details
References
[1] S. Shen-Orr, R. Milo, S. Mangan, U. Alon, "Network Motifs in the Transcriptional Regulation Network of Escherichia Coli," Nature Genetics, 31(1), 2002 pp. 64–68.
[2] R. Milo, S. Itzkovitz, N. Kashtan, R. Levitt, S. Shen–Orr, I. Ayzenshtat, M. Sheffer, and U. Alon, "Superfamilies of Designed and Evolved Networks," Science, 303(5663), 2004 pp. 1538–1542.
[3] H. Zenil and J. Tegnér, "Methods of Information Theory and Algorithmic Complexity for Network Biology," Biological Knowledge Discovery Handbook: Preprocessing, Mining and Postprocessing of Biological Data (M. Elloumi and A.Y. Zomaya, eds.), Hoboken, NJ: Wiley, forthcoming.
Permanent Citation
"Network Motifs and Graphlets"
http://demonstrations.wolfram.com/NetworkMotifsAndGraphlets/
Wolfram Demonstrations Project
Published: December 9 2013