Some Peptide Properties

Requires a Wolfram Notebook System
Interact on desktop, mobile and cloud with the free Wolfram Player or other Wolfram Language products.
All of a peptide's properties are determined by its amino acid sequence. This Demonstration shows the relationships between these properties and the sequence. The peptide mass, extinction coefficient, peptide charge and hydrophobic nature of the peptide are shown. Play around with the sequence of amino acids to see how the various properties of the peptide change, as shown both numerically and graphically on the various panels.
Contributed by: Alla Ahmad (May 2019)
Open content licensed under CC BY-NC-SA
Snapshots
Details
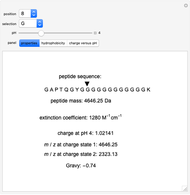
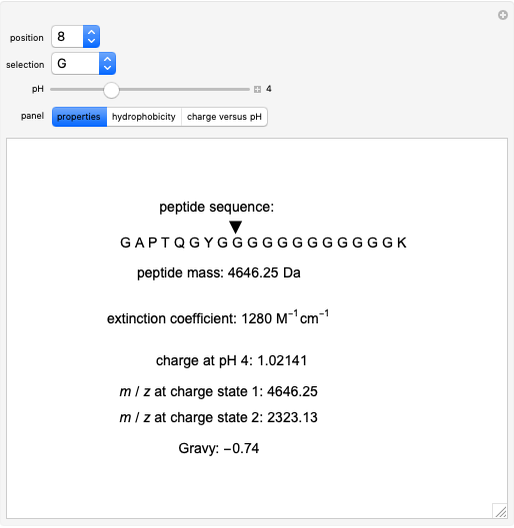
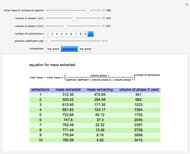
Peptide Mass
The peptide mass is calculated by adding the average masses for each amino acid in the sequence. A table of monoisotopic and average masses for all peptides is provided from the Mascot search engine website.
Extinction Coefficient
The extinction coefficient is principally dependent upon the number of tyrosines (Y) and tryptophans (W), but can also depend on the number of cysteines (C) if disulfide bonds are forming. The extinction coefficients for these amino acids and the appropriate formula can be found at Invitrogen's Peptide Calculator help page.
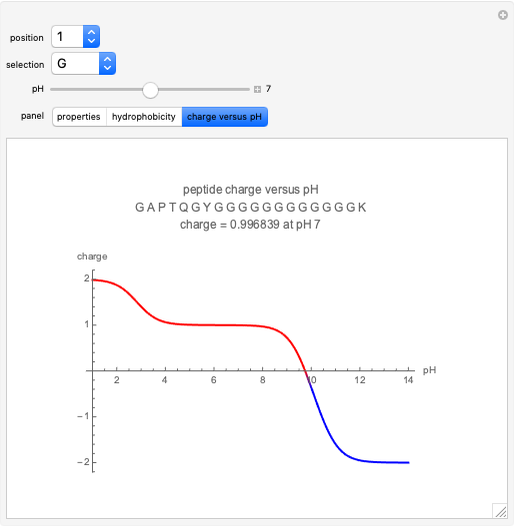
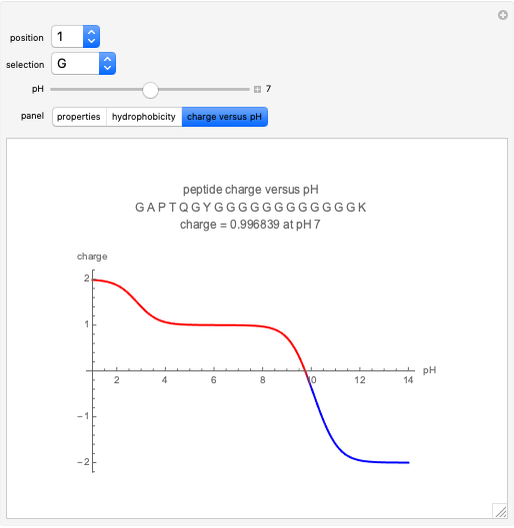
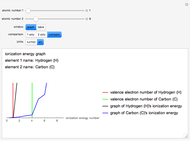
Charge
The charge of a peptide depends on the pH of the solution as well as the number of basic amino acids (lysine (K), arginine (R), histidine (H)), acidic amino acids (glutamic acid (E), aspartic acid (D)) on the N-terminal and C-terminal ends, and, to some extent, on the presence of cysteine (C) and tyrosine (Y). The values for pKas of specific side chains and the C/N termini can be found at the Sigma–Aldrich website. The equation for peptide charge can be found at Invitrogen's Peptide Calculator help page.
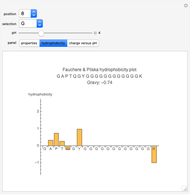
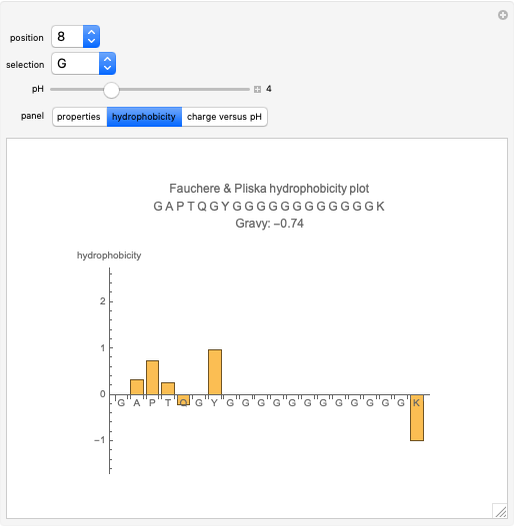
Hydrophobicity
Two different methods for hydrophobicity are used, one that is denoted Gravy and the other, Fauchere and Pliska. Gravy is calculated by taking the average of the hydrophobicity values of all the amino acids present in the peptide, as determined by Kyte and Doolittle [1] through protein structural analysis (negative is hydrophilic, positive is hydrophobic). The second method consists of graphing the hydrophobicity of each amino acid to show the trends of the peptide based on values determined by Fauchere and Pliska [2] for that amino acid's relative hydrophobicity compared to the amino acid glycine (negative is more hydrophilic than glycine, positive is more hydrophobic than glycine).
References
[1] J. Kyte and R. F. Doolittle, "A Simple Method for Displaying the Hydropathic Character of a Protein," Journal of Molecular Biology, 157(1), 1982 pp. 105–132. doi:10.1016/0022-2836(82)90515-0.
[2] J. L. Fauchere and V. Pliska. "Hydrophobic Parameters Pi of Amino-Acid Side Chains from the Partitioning of N-Acetyl-Amino-Acid Amides," European Journal of Medicinal Chemistry, 18(4), 1983 pp. 369–375. www.researchgate.net/publication/246404378_Hydrophobic _parameters _II _of _amino _acid _side-chains_from _the _partitioning _of _N-acetyl-amino_acid _amides.
Permanent Citation