Global and Local Sequence Alignment Algorithms

Requires a Wolfram Notebook System
Interact on desktop, mobile and cloud with the free Wolfram Player or other Wolfram Language products.
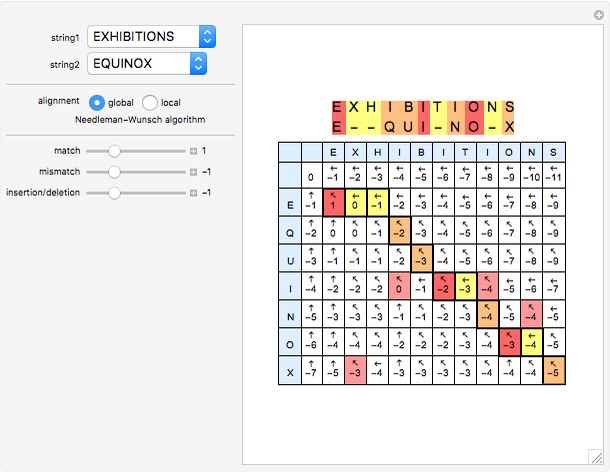
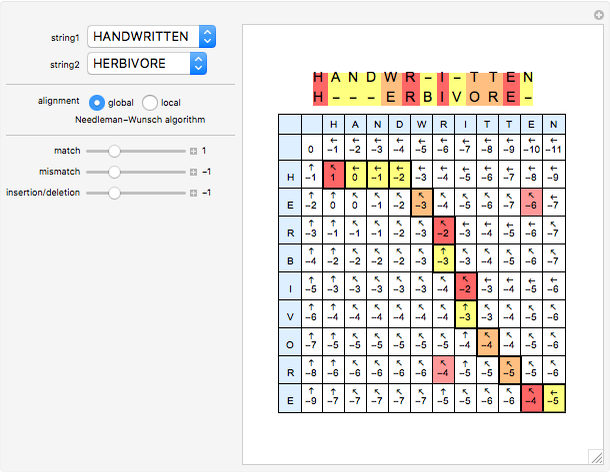
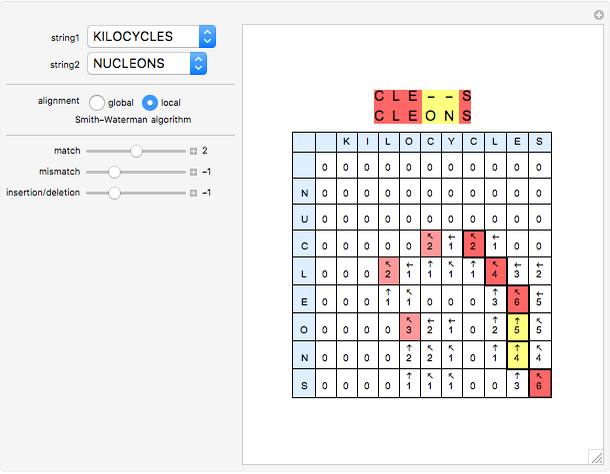
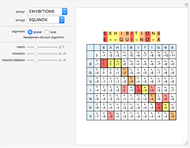
Sequence alignment is widely used in molecular biology to find similar DNA or protein sequences. These algorithms generally fall into two categories: global, which align the entire sequence, and local, which only look for highly similar subsequences. This Demonstration uses the Needleman–Wunsch (global) and Smith–Waterman (local) algorithms to align random English words. Gaps are shaded yellow, mismatches orange, and matches red (with a lighter shade for those matches not appearing in the final alignment).
Contributed by: Jordi Silvestre-Ryan (March 2011)
Open content licensed under CC BY-NC-SA
Snapshots
Details
This Demonstration uses a simple gap penalty, in which each insertion or deletion is scored the same. Because large gaps occur frequently in biological sequences, it is often better to use an affine gap penalty, which uses different values for opening and extending a gap.
For a technical description of the algorithms used, please see the following references:
N. C. Jones and P. A. Pevzner, An Introduction to Bioinformatics Algorithms, Cambridge, MA: The MIT Press, 2004.
I. Korf, M. Yandell, and J. Bedell, BLAST, Sebastopol, CA: O'Reilly & Associates Inc., 2003.
Permanent Citation
"Global and Local Sequence Alignment Algorithms"
http://demonstrations.wolfram.com/GlobalAndLocalSequenceAlignmentAlgorithms/
Wolfram Demonstrations Project
Published: March 7 2011