Handling Molecular Pathways in Systems Biology

Requires a Wolfram Notebook System
Interact on desktop, mobile and cloud with the free Wolfram Player or other Wolfram Language products.
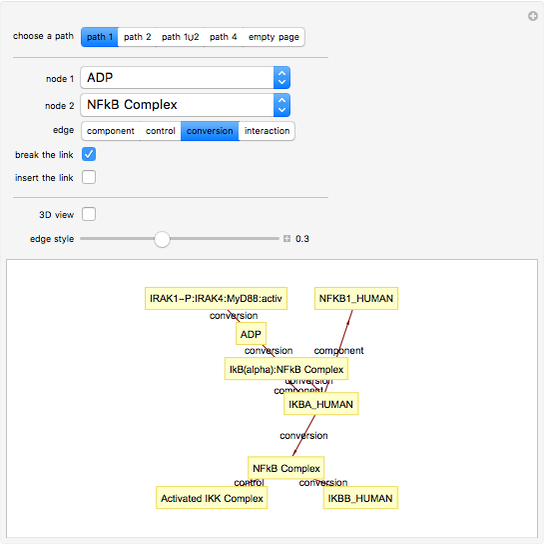
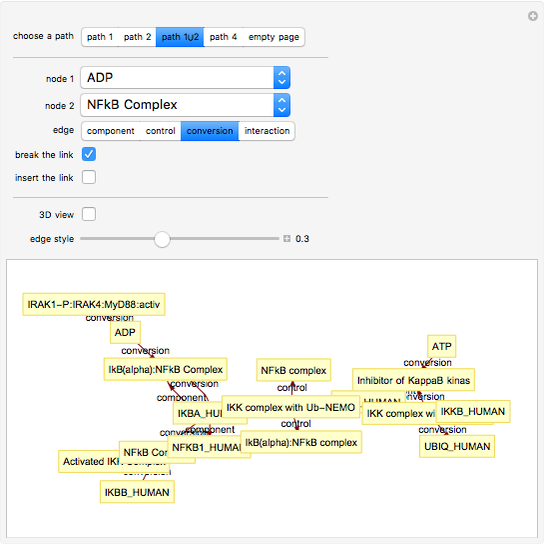
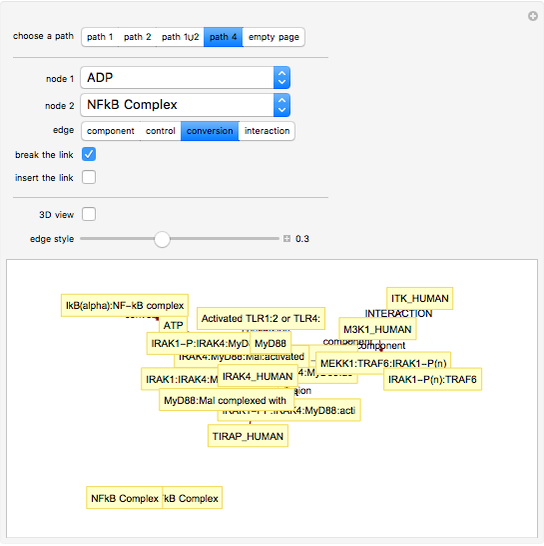
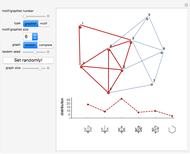
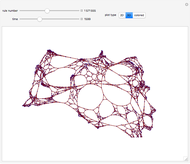
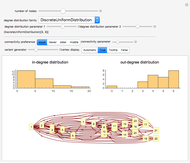
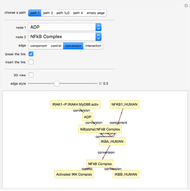
This Demonstration shows how to visualize and handle a pathway of proteins, such as the NF-kB pathway (nuclear factor-kappa B), split into three separated short paths: path 1, path 2, and path 4; path 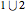 is a fusion of the first two pathways. The Demonstration does not discuss the biological implications of NF-kB. The only aim is to show how to use the Mathematica functions GraphPlot, Insert, and Delete to show a molecular pathway interactively. This could stimulate scientists and students to explore "visual" approaches to systems biology for other kinds of molecular mechanisms.
is a fusion of the first two pathways. The Demonstration does not discuss the biological implications of NF-kB. The only aim is to show how to use the Mathematica functions GraphPlot, Insert, and Delete to show a molecular pathway interactively. This could stimulate scientists and students to explore "visual" approaches to systems biology for other kinds of molecular mechanisms.
Contributed by: Luca Zammataro (March 2011)
Open content licensed under CC BY-NC-SA
Snapshots
Details
According to what we know about qualitative interaction among molecules, there are four kinds of relationships—component, control, conversion and interaction—that can occur between two proteins. Although there are limits on the number of proteins for the NF-kB pathway proposed here, there are no limits on the addition of new pathways. Also, you can remove an existing pathway and observe how the network behavior changes after that specific junction among two or more nodes has been removed. You can also create new pathways starting from an empty page, filling it by means of the "nodes" and "edges" controls.
The NF-kB pathway has been obtained by means of cytoscape software: http://www.cytoscape.org/
Permanent Citation
"Handling Molecular Pathways in Systems Biology"
http://demonstrations.wolfram.com/HandlingMolecularPathwaysInSystemsBiology/
Wolfram Demonstrations Project
Published: March 7 2011