Uncertainties in Isothermal Microbial Growth

Requires a Wolfram Notebook System
Interact on desktop, mobile and cloud with the free Wolfram Player or other Wolfram Language products.
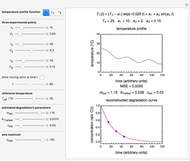
This Demonstration estimates a theoretical spread in the size of a microbial population in a closed habitat at a constant temperature at a given time. It is based on the assumption that the growth curve follows the shifted logistic model. Hence the growth ratio, linear or logarithmic, is determined by three parameters: the approximate asymptotic value  , a measure of the growth curve's steepness at the exponential phase
, a measure of the growth curve's steepness at the exponential phase  , and a marker of the inflection point
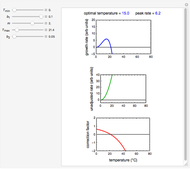
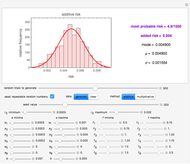
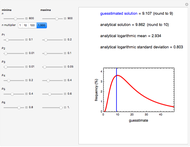
, and a marker of the inflection point 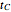 . The three parameters are entered as the lower and upper limits of their plausible ranges. Monte Carlo simulations generate numerous hypothetical growth curves with random parameter values within the chosen ranges. The Demonstration displays a representative growth curve for the chosen parameter settings and the corresponding histogram and calculates the most probable population size for the chosen time along with some statistical characteristics of the generated data.
. The three parameters are entered as the lower and upper limits of their plausible ranges. Monte Carlo simulations generate numerous hypothetical growth curves with random parameter values within the chosen ranges. The Demonstration displays a representative growth curve for the chosen parameter settings and the corresponding histogram and calculates the most probable population size for the chosen time along with some statistical characteristics of the generated data.
Contributed by: Mark D. Normand and Micha Peleg (December 2011)
Open content licensed under CC BY-NC-SA
Snapshots
Details
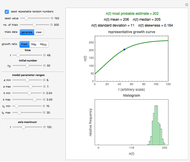
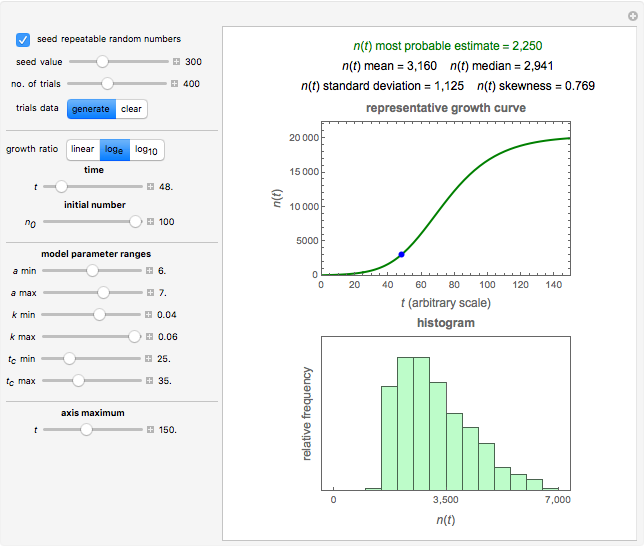
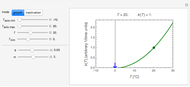
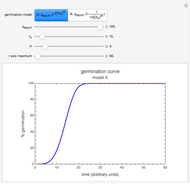
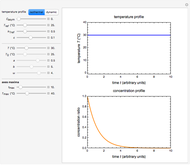
Snapshot 1: growth curve of 50 cells after 48 time units and corresponding histogram of the generated numbers calculated using the linear growth ratio
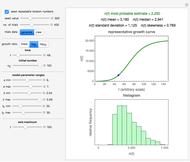
Snapshot 2: growth curve of 100 cells after 48 time units and corresponding histogram of the generated numbers calculated using the natural logarithmic growth ratio
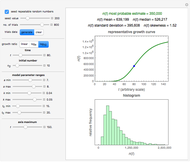
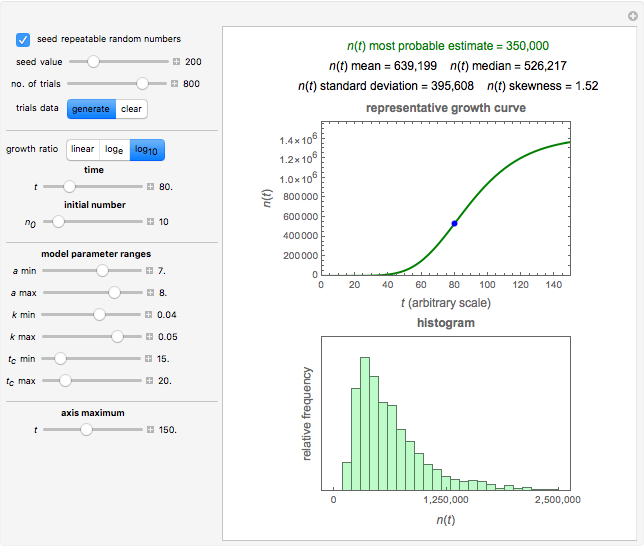
Snapshot 3: growth curve of 10 cells after 80 time units and corresponding histogram of the generated numbers calculated using the base 10 logarithmic growth ratio
The isothermal growth curve of a microorganism is a plot of its population size (usually expressed as a count) versus time. When the growth takes place in a closed habitat and at a constant temperature, the curve is typically sigmoid initially and consists of three regions representing a "lag phase", an "exponential growth phase", and a "stationary phase". However, the latter cannot last indefinitely and eventually ends up in the onset of mortality. The sigmoid part of the growth curve can be described by a variety of interchangeable growth models, the shifted logistic model among them [1]. For mild, moderate, and intensive growth this model can be written as 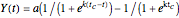 . For mild growth,
. For mild growth, 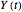 might be the linear growth ratio
might be the linear growth ratio 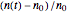 , for moderate growth the natural logarithmic ratio
, for moderate growth the natural logarithmic ratio 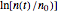 , and for intensive growth, where the population rises by several orders of magnitude,
, and for intensive growth, where the population rises by several orders of magnitude, 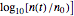 . In all three cases,
. In all three cases, 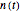 is the temporary number of cells after time
is the temporary number of cells after time  and
and 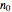 their initial number. This implies that the growth curve itself in the three cases will be expressed as
their initial number. This implies that the growth curve itself in the three cases will be expressed as 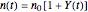 ,
, 
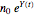 , or
, or 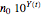 , respectively.
, respectively.
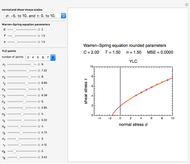
According to this model, the growth parameters are  (dimensionless), approximately the asymptotic growth ratio's level at the stationary phase,
(dimensionless), approximately the asymptotic growth ratio's level at the stationary phase,  (time units reciprocal), a measure of the growth curve's steepness at its inflection point (the actual slope is
(time units reciprocal), a measure of the growth curve's steepness at its inflection point (the actual slope is 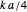 ), and
), and 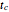 (time units), a marker of the inflection point.
(time units), a marker of the inflection point.
Since foods' composition and physical properties can and frequently do vary, there is always a degree of uncertainty concerning the magnitude of experimentally determined microbial growth parameters, and consequently in the predicted population size.
In this Demonstration you can enter the time  , the plausible lower and upper bounds of the shifted logistic model's growth parameters,
, the plausible lower and upper bounds of the shifted logistic model's growth parameters, 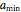 ,
, 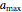 ,
, 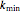 ,
, 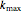 ,
,  , and
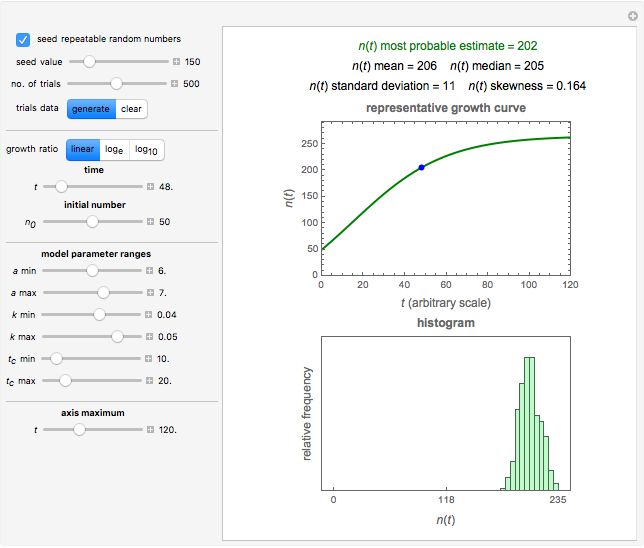
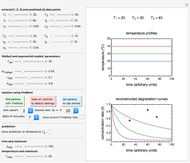
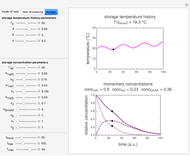
, and 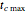 with sliders, and also choose the number of Monte Carlo trials. Clicking the "seed repeatable random numbers" checkbox lets you reproduce the random simulation from the same seed value, also entered with a slider. The program then generates the chosen number of trials with random combinations of the growth parameters, assumed to be uniformly distributed within their respective ranges ("maximum ignorance") and calculates the corresponding growth curve using the shifted logistic model's equation with the chosen definition of the growth ratio (linear, natural logarithmic, or base 10 logarithmic) entered with a setter. The program plots a representative growth curve for the chosen parameters on which the slider-entered time
with sliders, and also choose the number of Monte Carlo trials. Clicking the "seed repeatable random numbers" checkbox lets you reproduce the random simulation from the same seed value, also entered with a slider. The program then generates the chosen number of trials with random combinations of the growth parameters, assumed to be uniformly distributed within their respective ranges ("maximum ignorance") and calculates the corresponding growth curve using the shifted logistic model's equation with the chosen definition of the growth ratio (linear, natural logarithmic, or base 10 logarithmic) entered with a setter. The program plots a representative growth curve for the chosen parameters on which the slider-entered time  is shown as a moving blue dot, and below it the histogram of the generated
is shown as a moving blue dot, and below it the histogram of the generated 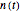 s for the entered settings. The most probable estimate of
s for the entered settings. The most probable estimate of 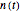 is the mean value of the histogram's bin having the largest number of entries. It is displayed accompanied by the generated counts' mean, median, standard deviation, and skewness. In order to produce new random trial data with exactly the same parameter settings, first click the "clear" and then the "generate" buttons.
is the mean value of the histogram's bin having the largest number of entries. It is displayed accompanied by the generated counts' mean, median, standard deviation, and skewness. In order to produce new random trial data with exactly the same parameter settings, first click the "clear" and then the "generate" buttons.
Not all possible control settings produce realistic survival curves when the arbitrary time units are replaced by hours or days. The initial number's allowed range is 1 to 100. For larger numbers, multiply the results rendered by the Demonstration by the appropriate factor.
References
[1] M. Peleg, Advanced Quantitative Microbiology for Foods and Biosystems: Models for Predicting Growth and Inactivation, Boca Raton, FL: CRC Press, 2006.
[2] M. A. J. S. von Boekel, Kinetic Modeling of Reactions in Foods, Boca Raton: CRC Press, 2008.
[3] M. Peleg and M. G. Corradini, "Microbial Growth Curves — What the Models Tell Us and What They Cannot," CRC Critical Reviews in Food Science, 51, 2011 pp. 917–945.
[4] S. Smith-Simpson, M. G. Corradini, M. D. Normand, M. Peleg, and D. W. Schaffner, "Estimating Microbial Growth Parameters from Non-Isothermal Data: A Case Study with Clostridium perfringens," International Journal of Food Microbiology, 118, 2007 pp. 294–303.
[5] M. G. Corradini, A. Amézquita, M. D. Normand, and M. Peleg, "Modeling and Predicting Non-isothermal Microbial Growth Using General Purpose Software," International Journal of Food Microbiology, 106, 2006 pp. 223–328.
Permanent Citation